Topic 2: Crystal Structure Analysis of Macromolecular Complex
Elucidation of Ultimate Switching Mechanisms Hidden in Protein Molecules
Bacteria such as Salmonella and Escherichia coli move by rotating several flagella, which are similar to propellers. The flagellum comprises a rotary motor, bearing, and a rotating shaft at the base. The flagellar filaments rotate at 200-400 revolutions per second, like a propeller, allowing bacteria to move a distance 20 times larger than their body size each second. In humans, this would correspond to a speed of 100 km/hour. The flagellum is a powerful yet delicate nanomachine. The switching mechanisms of constituent protein molecules also enable bacteria to quickly change direction. At SPring-8, we have elucidated, for the first time, the switching mechanisms of the flagella, which are the ultimate energy-efficient ecological machine and can provide many clues to developing nanotechnology.
Crystal Structure Analyses of Proteins at the Atomic Level
A flagellar filament is comprised of a cylindrical bundle of eleven long, thin protofilaments, which are 20 nm (1 nm = 10-9 m) in diameter and 10-15 μm (1 μm = 10-6 m) in length (Fig. 1). Each protofilament is an assembly of a single protein, flagellin, with a molecular weight of about 50 kDa1) . If all the flagellin molecules were to have equal size and shape, flagella filaments would only assume a linear conformation, and no propelling power would be produced even if the flagellar motor is rotating at high speed. However, bacteria can move freely and change direction quickly. This is achieved via the conformation of the left-handed helical structure of flagella, which are slightly twisted due to the existence of protofilaments with slightly different sizes (5.19 nm and 5.27 nm in the intersubunit distance).
When a flagellar motor rotating at high speed inside a cell membrane suddenly reverses its direction, flagella filaments are twisted, and the structures of several protofilaments are modified to change the helix from left-handed to right-handed. Then, flagella that are bundled together when moving straight are unraveled, and the propelling force becomes unbalanced; this results in a change in the direction of movement. This quick directional change is enabled by a very small, sub-nanoscale difference (0.08 nm) in the intersubunit distances of flagelin.
Many scientists around the world who aim to develop nanomachines have been intensely studying flagella. The secondary structure distributions and alignment patterns of flagellar filaments had been elucidated, but the high-precision switching mechanisms remained to be identified. To clarify the mechanisms, detailed structure determination was required.
In August 1996, Dr. Kei-ichi Namba2) (the research director of the Advance Technology Research Laboratories, Matsushita Electric Industrial Co., Ltd., Japan.) led the Namba Protonic Nanomachine Project3) to explore the frontiers of flagellar research. “We tried to elucidate the 3D structures of flagellin molecules but the size of the flagellin crystal samples was at most a few micrometers, with which we had no hope of obtaining reliable data using conventional X-ray sources at that time,” recalled Dr. Namba, looking back on that time.
Then, they decided to use highly brilliant X-rays at SPring-8, which offered the world best analyzing power, for their experiments.
However, they faced another difficulty. Determination of the 3D structures of flagellin molecules requires crystal molecules to be perfectly aligned. Crystallization of flagellin molecules was difficult, since they tend to be polymerized in a long fibrous form. To overcome this difficulty, they removed the domains important for fiber formation and structure stabilization from flagellin molecules of Salmonella flagella. Removal of these domains yielded a 41-kDa fragment (F41) that lacked fiber formation capability. Using this fragment, they succeeded in preparing high-quality crystal samples.
In February 1998, the researchers started experiments on these samples, applying the multi-wavelength anomalous diffraction (MAD) technique at the RIKEN Structural Biology I beamline (BL45XU). MAD is a method of analyzing crystal structures, based on the fact that the strength of X-rays scattered off of a specific type of atoms in protein crystal samples depends on the wavelength of irradiated X-rays. “We faced crushing difficulties in obtaining good data from these crystal samples, since they were in thin plate-shape and only a few micrometers in thickness. After undertaking tireless efforts to keep the samples flat after freezing, we finally determined the structure of flagellin molecules with 0.2-nm resolution,” recalled Dr. Namba.
1) 103 Da (molecular mass unit)
2) Currently Professor at Osaka University, Japan.
3) The Exploratory Research for Advanced Technology (ERATO), Japan Science and Technology Agency.
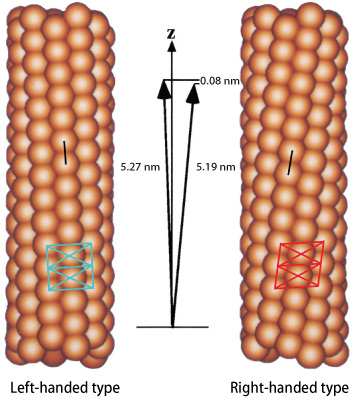
Flagellin molecules are depicted in particle-shape. The intersubunit distance of protofilaments is ‾5.2 nm. However, that of the right-handed form and that of the left-handed form differ by 0.08 nm. This difference enables the conformation of a curved and twisted helical propeller.
Reproduction of Switching Actions using Computer Simulation
F41 consists of three domains: D1, D2, and D3. Domain D1 comprises three long α-helices and a β-hairpin. An α-helix has a structure in which the polypeptide backbone (the basic building block of proteins) are folded in right-handed helical form, whereas a β-hairpin has a structure in which the polypeptide backbone is sharply bent, like a hairpin. Domains D2 and D3 mostly comprise β-sheets and β-hairpins. Surprisingly, the conformation of flagellar protofilaments could be observed in the crystal, which had not been expected because the monomers are crystallized. The protofilament forms a sheet structure by alternatively overlapping to conform with the shape of the crystal.
Additionally, this research group performed computer simulation studies on the behavior of flagellin molecules, in order to examine the switching structures of protofilaments in further detail. A computer model of protofilaments consisting of three flagellin molecules was constructed. The bottom molecule was pulled down step-by-step in 0.01-nm increments, with the top molecule staying fixed, in order to observe structural deformations of the middle molecule under tension. In the beginning, the middle flagellin molecule was only stretched gradually. However, at a certain point, β-hairpins in the areas where two molecules contact each other changed their structure slightly but suddenly (Fig. 2). “This finding indicates that there is a switch that is responsible for changing the intersubunit distance by 0.08 nm in the structure near the hairpins,” said Dr. Namba. This is exactly the moment when the “0.08 nm shift” of flagellin molecules was revealed at SPring-8. Thus, Dr. Namba and colleagues had finally discovered the sophisticated mechanical switch hidden in flagellin molecules.
“Flagellin molecules are not special proteins. In our study, we demonstrated one example of a sophisticated action mechanism by which many proteins function,” said Dr. Namba.
Nanotechnology researchers are investigating the structures and functions of proteins, since proteins behave as nanomachines and share many design principles. Therefore, the amazing mechanisms of proteins serve as a model for artificial nanomachines. By elucidating the 3D structures of proteins that function as nanomachines, we will better understand the design principles of highly flexible and sophisticated nanomachines. Analysis technologies offered by SPring-8 are driving extensive exploration of the nanoscale world.
This achievement of the ERATO Namba Protonic Nanomachine Project that elucidated the high-precision switching mechanisms of protein molecules was published in Nature (March 2001). The 3D structure of rotating flagellar filaments appeared on the journal's cover.
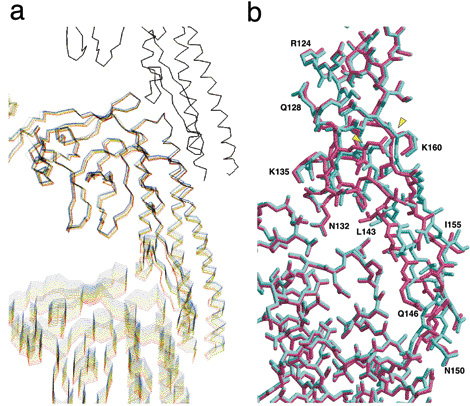
a. Computer simulation of a protofilament consisting of three flagellin molecules. Structure changes of flagellin molecules, which contribute the high precision switching that changes the intersubunit distance of protofilaments, are depicted. The bottom molecule is gradually pulled down at 0.01-nm intervals, while the top molecule remains fixed. This process is intended to stretch the protofilament and add mechanical stress to the middle molecule. Simulated images of twelve steps are overlaid every 0.05 nm by color-coding these steps with rainbow colors ranging from blue to red. Entire molecules are gradually stretched in the first nine steps (total stretch of 0.45 nm) but clear structural changes around β-hairpins can be observed between steps 9 and 10 (total stretch of 0.5 nm).
b. Magnified views of β-hairpins and surrounding areas where structural changes are observed. Two images before (cyan) and after (dark pink) the switching are overlaid. Only the β-hairpins (a structure stretching from middle right toward bottom right; from N132 to K160 via L143, Q146, N150, and I155) of the upper flagellin molecule show sudden structure changes, suggesting the existence of switching actions in this area.
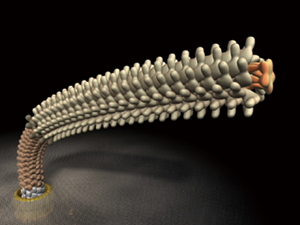
A flagellum consists of ring and helix structures in which ‾30 kinds of constituent protein molecules self-assemble to form a cluster. The number of molecules in each cluster ranges from several for the smallest to tens of thousands for the largest. The well-organized self-assembly process of these structures also provides clues that will be invaluable in the development of nanomachines.
Reference
1. F. A. Samatey, K. Imada, S. Nagashima, F. Vonderviszt, T. Kumasaka, M. Yamamoto and K. Namba; Nature, 410, 331 (2001)